

细菌转录翻译偶联机制研究
Recent advances of transcription-translation coupling in bacteria
Received date: 2024-03-11
Online published: 2024-04-19
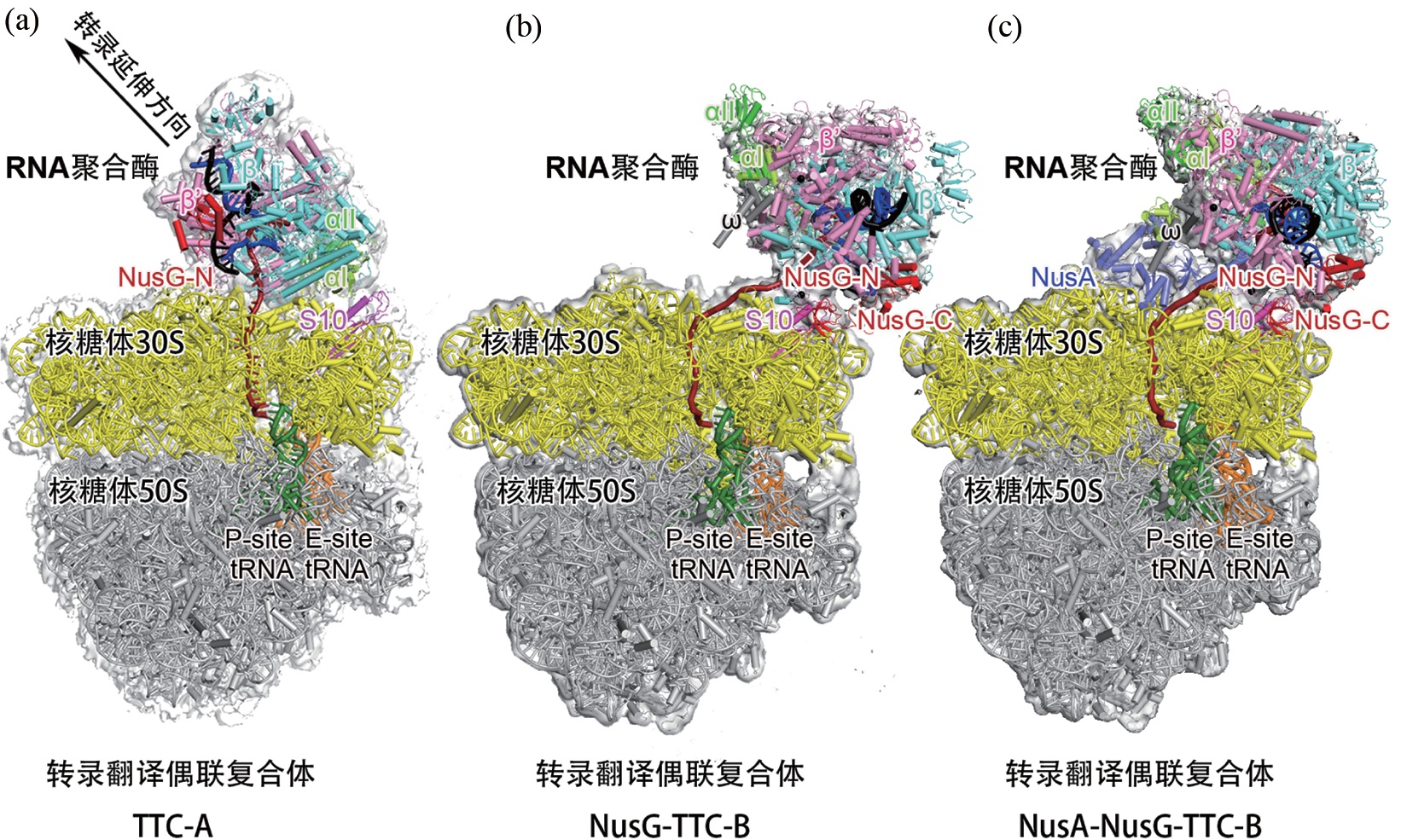
张晶, 王程远 . 细菌转录翻译偶联机制研究[J]. 自然杂志, 2024 , 46(2) : 95 -104 . DOI: 10.3969/j.issn.0253-9608.2024.02.003

[16] GUO X, MYASNIKOV A G, CHEN J, et al. Structural basis for NusA stabilized transcriptional pausing [J]. Mol Cell, 2018, 69(5):
816-827.e4.
[17] NUDLER E, MUSTAEV A, GOLDFARB A, et al. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase [J]. Cell, 1997, 89(1): 33-41.
[18] KOMISSAROVA N, KASHLEV M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3´ end of the RNA intact and extruded [J]. Proc Natl Acad Sci USA, 1997, 94(5): 1755-1760.
[19] JU X, LI S, FROOM R, et al. Incomplete transcripts dominate the Mycobacterium tuberculosis transcriptome [J]. Nature, 2024,
627(8003): 424-430.
[20] PROSHKIN S, RAHMOUNI A R, MIRONOV A, et al. Cooperation between translating ribosomes and RNA polymerase in transcription elongation [J]. Science, 2010, 328(5977): 504-508.
[21] WEBSTER M W, WEIXLBAUMER A. Macromolecular assemblies supporting transcription-translation coupling [J]. Transcription,
2021, 12(4): 103-125.
[22] WEE L M, TONG A B, FLOREZ ARIZA A J, et al. A trailing ribosome speeds up RNA polymerase at the expense of transcript
fidelity via force and allostery [J]. Cell, 2023, 186(6): 1244-1262.e34.
[23] JACQUET M, KEPES A. Initiation, elongation and inactivation of lac messenger RNA in Escherichia coli studied by measurement
of its β-galactosidase synthesizing capacity in vivo [J]. J Mol Biol, 1971, 60(3): 453-472.
[24] GOURSE R L, CHEN A Y, GOPALKRISHNAN S, et al. Transcriptional responses to ppGpp and DksA [J]. Annu Rev Microbiol, 2018, 72: 163-184.
[25] MOLODTSOV V, SINEVA E, ZHANG L, et al. Allosteric effector ppGpp potentiates the inhibition of transcript initiation by DksA [J].
Mol Cell, 2018, 69(5): 828-839.e5.
[26] SANCHEZ-VAZQUEZ P, DEWEY C N, KITTEN N, et al. Genome-wide effects on Escherichia coli transcription from ppGpp
binding to its two sites on RNA polymerase [J]. Proc Natl Acad Sci USA, 2019, 116(17): 8310-8319.
[27] WANG B, ARTSIMOVITCH I. NusG, an ancient yet rapidly evolving transcription factor [J]. Front Microbiol, 2021, 11: 619618.
[28] SULLIVAN S L, WARD D F, GOTTESMAN M E. Effect of Escherichia coli NusG function on lambda N-mediated transcription antitermination [J]. J Bacteriol, 1992, 174(4): 1339-1344.
[29] MOONEY R A, SCHWEIMER K, RÖSCH P, et al. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators [J]. J Mol Biol, 2009, 391(2): 341-358.
[30] BURMANN B, KNAUER S, SEVOSTYANOVA A, et al. An α helix to β barrel domain switch transforms the transcription factor
RfaH into a translation factor [J]. Cell, 2012, 150(2): 291-303.
[31] MCGARY K, NUDLER E. RNA polymerase and the ribosome: the close relationship [J]. Curr Opin Microbiol, 2013, 16(2): 112-117.
[32] PAN T, ARTSIMOVITCH I, FANG X W, et al. Folding of a large ribozyme during transcription and the effect of the elongation factor
NusA [J]. Proc Natl Acad Sci USA, 1999, 96(17): 9545-9550.
[33] STRAUß M, VITIELLO C, SCHWEIMER K, et al. Transcription is regulated by NusA: NusG interaction [J]. Nucleic Acids Res, 2016,
44(12): 5971-5982.
[34] MANDELL Z F, OSHIRO R T, YAKHNIN A V, et al. NusG is an intrinsic transcription termination factor that stimulates motility and
coordinates gene expression with NusA [J]. eLife, 2021, 10: e61880.
[35] ARTSIMOVITCH I, LANDICK R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals
[J]. Proc Natl Acad Sci USA, 2000, 97(13): 7090-7095.
[36] MITRA P, GHOSH G, HAFEEZUNNISA M, et al. Rho protein: roles and mechanisms [J]. Annu Rev Microbiol, 2017, 71: 687-709.
[37] KRUPP F, SAID N, HUANG Y H, et al. Structural basis for the action of an all-purpose transcription anti-termination factor [J]. Mol
Cell, 2019, 74(1): 143-157.e5.
[38] WANG C, MOLODTSOV V, FIRLAR E, et al. Structural basis of transcription-translation coupling [J]. Science, 2020, 369(6509): 1359-1365.
[39] KOHLER R, MOONEY R A, MILLS D J, et al. Architecture of a transcribing-translating expressome [J]. Science, 2017, 356(6334):
194-197.
[40] KANG J Y, MOONEY R A, NEDIALKOV Y, et al. Structural basis for transcript elongation control by NusG family universal regulators [J]. Cell, 2018, 173(7): 1650-1662.e14.
[41] MOLODTSOV V, WANG C, KAELBER J T, et al. Structural basis of RfaH-mediated transcription-translation coupling [J/OL]. [2024-
03-11]. https://www.biorxiv.org/content/10.1101/2023.11.05.565726v1.full.pdf.
[42] ZUBER P K, ARTSIMOVITCH I, NANDYMAZUMDAR M, et al. The universally-conserved transcription factor RfaH is recruited to a
hairpin structure of the non-template DNA strand [J]. eLife, 2018, 7: 36349.
[43] FAN H, CONN A B, WILLIAMS P B, et al. Transcription–translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits [J]. Nucleic Acids Res, 2017, 45(19): 11043-11055.
[44] DEMO G, RASOULY A, VASILYEV N, et al. Structure of RNA polymerase bound to ribosomal 30S subunit [J]. eLife, 2017, 6: e28560.
[45] WEBSTER M W, TAKACS M, ZHU C, et al. Structural basis of transcription-translation coupling and collision in bacteria [J]. Science, 2020, 369(6509): 1355-1359.
[46] O´REILLY F J, XUE L, GRAZIADEI A, et al. In-cell architecture of an actively transcribing-translating expressome [J]. Science, 2020, 369(6503): 554-557.
[47] JOHNSON G E, LALANNE J B, PETERS M L, et al. Functionally uncoupled transcription-translation in Bacillus subtilis [J]. Nature,
2020, 585(7823): 124-128.
[48] XIONG H B, PAN H M, LONG Q Y, et al. AtNusG, a chloroplast nucleoid protein of bacterial origin linking chloroplast transcriptional and translational machineries, is required for proper chloroplast gene expression in Arabidopsis thaliana [J]. Nucleic Acids Res, 2022, 50(12): 6715-6734.
/
| 〈 |
|
〉 |